To Determine the Order of Genes on a Chromosome, One Can Perform a
How would you feel if you had to be the ane to challenge Gregor Mendel'southward paradigm-shifting laws of inheritance? Withal Thomas Hunt Morgan did exactly this and in the process fabricated gene mapping possible.
In 1911, while studying the chromosome theory of heredity, biologist Thomas Hunt Morgan had a major breakthrough. Morgan occasionally noticed that "linked" traits would separate. Meanwhile, other traits on the same chromosome showed trivial detectable linkage. Morgan considered the prove and proposed that a procedure of crossing over, or recombination, might explain his results. Specifically, he proposed that the two paired chromosomes could "cross over" to commutation data. Today, we know that recombination does indeed occur during prophase of meiosis (Figure 1), and information technology creates unlike combinations of alleles in the gametes that event (i.e., the F1 generation; Figure two).
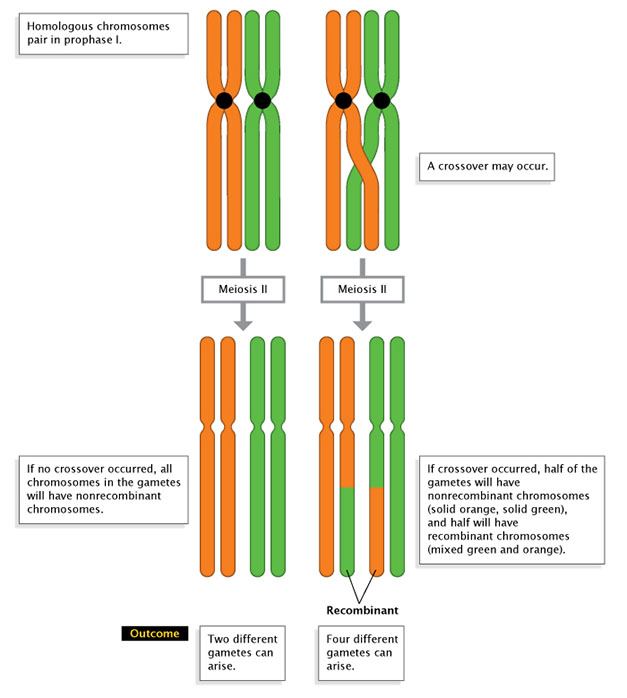
Effigy ane: Recombination and gamete product.
A comparing of nonrecombination (left) with recombination (right), shows how recombination affects the way chromosomes are passed into gametes in Meiosis II. On the right, a single crossover event produces one-half nonrecombinant gametes and half recombinant gametes.
© 2022 Nature Education Adjusted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2d ed. All rights reserved. ![]()
When proposing the idea of crossing over, Morgan besides hypothesized that the frequency of recombination was related to the distance between the genes on a chromosome, and that the interchange of genetic information bankrupt the linkage between genes. Morgan imagined that genes on chromosomes were similar to pearls on a string (Weiner, 1999); in other words, they were physical objects. The closer 2 genes were to one some other on a chromosome, the greater their chance of beingness inherited together. In dissimilarity, genes located farther abroad from one some other on the same chromosome were more likely to be separated during recombination. Therefore, Morgan correctly proposed that the strength of linkage betwixt ii genes depends upon the altitude between the genes on the chromosome. This proffer became the basis for construction of the earliest maps of the human genome.
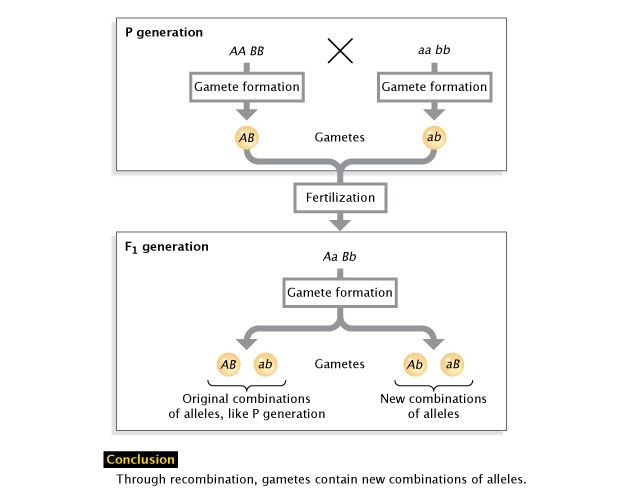
Effigy 2: Allele recombination.
Recombination is the sorting of alleles into new combinations. Following the germination of gametes over two generations shows how recombination tin produce new allelic combinations (lower right) or stay the aforementioned (lower left).
© 2022 Nature Educational activity Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. ![]()
Sturtevant Uses Crossing-Over Data to Construct the First Genetic Map
Soon afterward Morgan presented his hypothesis, Alfred Henry Sturtevant, a nineteen-yr-old Columbia University undergraduate who was working with Morgan, realized that if the frequency of crossing over was related to distance, i could use this information to map out the genes on a chromosome. After all, the further apart two genes were on a chromosome, the more likely information technology was that these genes would dissever during recombination. Therefore, as Sturtevant explained it, the "proportion of crossovers could be used every bit an index of the altitude between whatsoever two factors" (Sturtevant, 1913). Collecting a stack of laboratory data, Sturtevant went home and spent about of the night drawing the start chromosomal linkage map for the genes located on the X chromosome of fruit flies (Weiner, 1999).
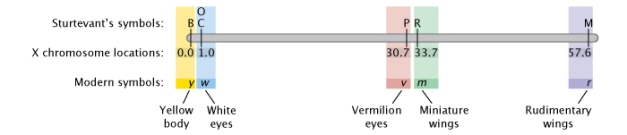
Effigy iii: Sturtevant'southward Drosophila gene map.
In Sturtevant's gene map, six traits are bundled along a linear chromosome co-ordinate to the relative altitude of each from trait B. Traits include yellow body (B), white optics (C, O), Vermillion eyes (P), miniature wings (R), and rudimentary wings (One thousand).
© 2022 Nature Didactics Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2d ed. All rights reserved. ![]()
When creating his map, Sturtevant started past placing half dozen X-linked genes in club. B was a factor for black body color. C was a factor that immune color to appear in the optics. Flies with the P gene had vermilion eyes instead of the ordinary ruby-red, and flies with 2 copies of the recessive O gene had eyes that appeared a shade known every bit eosin. The R and M factors both afflicted the wings. Sturtevant placed C and O at the same point because they were completely linked and were e'er inherited together — in other words, he never saw any evidence for recombination between C and O. Sturtevant then placed the remainder of the genes in the lodge shown in Figure 3 (Sturtevant, 1913). Crossover events were tracked by examining the F2 progeny in crosses for "new" phenotypes.

Figure 4: Phenotypes used in Sturtevant'south cross.
Sturtevant crossed flies with long wings (Chiliad) and vermillion optics (p) with flies with rudimentary wings (m) and red eyes (P). These traits are X-linked.
For case, to detect the distance between P (vermilion eyes) and M (long wings), Sturtevant performed crosses between flies that had long wings and vermilion eyes and flies that had modest wings and red optics. These crosses resulted in F1 flies that either had long wings and red optics or long wings and vermilion eyes. Sturtevant then crossed these 2 types of F1 flies and analyzed the offspring for evidence of recombination. Unexpected phenotypes observed in the male F2 progeny from this cross were then examined. (Because very little recombination occurs in the male germ line of Drosophila, only the female F1 chromosomes are considered for predicting phenotypes [Figure iv].) Sturtevant noted four classes of male flies in this Fii generation, as shown in Table 1.
The 2 additional classes of flies that appeared in this generation (long wings with red eyes and rudimentary wings with vermilion eyes) could but be explained by recombination occurring in the female germ line.
| Phenotype | Number of Flies | Nature of Related Gametes |
| Long wings, red optics | 105 | Recombinant |
| Rudimentary wings, cherry-red eyes | 33 | Nonrecombinant |
| Long wings, vermilion optics | 316 | Nonrecombinant |
| Rudimentary wings, vermilion optics | iv | Recombinant |
| Tabular array 1: Class of male files in the Ftwo generation | ||
Sturtevant then worked out the order and the linear distances between these linked genes, thus forming a linkage map. In doing and then, he computed the altitude in an arbitrary unit he called the "map unit," which represented a recombination frequency of 0.01, or one%. Later, the map unit was renamed the centimorgan (cM), in honor of Thomas Chase Morgan, and it is still used today as the unit of distances along chromosomes.
In add-on to describing the lodge of the genes on the X chromosome of fruit flies, Sturtevant'southward 1913 paper elucidated a number of other interesting points, including the following:
- The human relationship between crossing over and genetic map distance
- The effects of multiple crossover events
- The fact that a first crossover can inhibit a 2d crossover (a phenomenon called interference, which is described afterwards in this article)
Mapping Genes Using Recombination Frequency
To ameliorate understand how Sturtevant arrived as his results, let's have a closer look at the process he followed. In Figure 5, the gray-eosin and yellow-cherry-red flies are the parental lines, and all the alleles for these traits are linked on the 10 chromosome. Therefore, any gray-red or yellow-eosin male offspring are recombinants. As you tin can see, two recombinants issue from the cross. We count only the male progeny because the males have one X chromosome and dominance will not obscure any phenotypes (Robbins, 2000). Of course, crossing over tin occur only in the female fruit flies, which have two 10 chromosomes. Thus, in this cantankerous, the female person F1 gametes provide the parental and recombinant gametes that we detect in the Ftwo progeny.
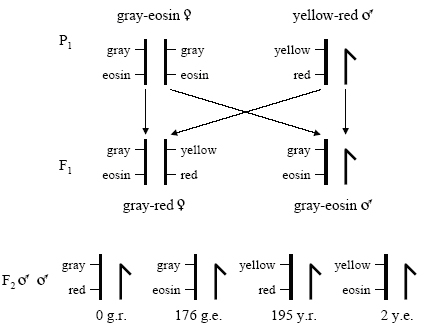
Figure 5: An analogy of Sturtevant's cross, showing the chromosomes, illustrates his logic
Sturtevant illustrated the crosses and offspring resulting from a parental strain of grayness-eosin female flies and yellow-crimson male flies. He followed this cantankerous to the F2 generation.
© 2008 Robert J. Robbins. All rights reserved. ![]()
In social club to calculate the recombination frequency nosotros use the post-obit formula:
Substituting the values from our data fix, nosotros arrive at the post-obit:
Therefore, the two genes are 0.5 map units apart.
Deviations from Expected Results Revealed Genetic Interference
Sturtevant as well described the fact that, for genes that were distant from ane some other, there was a discrepancy in the predicted number of crossovers. For example, the distance between B and Yard on his map was 57.6. His recombination data using those two genes, however, did non suggest this altitude. Instead, Sturtevant constitute 260 recombinants in 693 male progeny, which, when plugged into the equation, produced a result of 37.six. How, so, did Sturtevant explain the deviation?
In short, Sturtevant realized that double recombination events could occur if genes were far apart. Moreover, non only did Sturtevant's data suggest that double-crossing over occurred, but it besides suggested that an initial crossover event could inhibit subsequent events by mode of a miracle Sturtevant referred to every bit interference.
To understand how Sturtevant arrived at this conclusion, take a look at the information shown in Figure 6 (Sturtevant, 1913). As you can see, Sturtevant examined recombination events betwixt B (body colour), CO (two eye color genes that were closely linked), and R (rudimentary wings), and compared the frequencies of crossover events. When B and CO did non split, Sturtevant noticed that the "gametic ratio," or presence, of CO/R recombinants was approximately 1:two (3,454:6,972). However, when a crossover between B/CO (N = sixty) occurred, there was a much lower likelihood (approximately 1:6.five) of a crossover between CO/R (N = 9). This finding is indicative of interference.
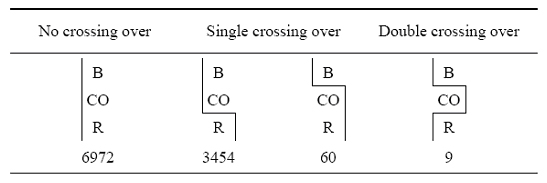
Figure six: Data collected past Sturtevant
Number of possible combinations in forms having from ii to 36 chromosomes in the pre-synaptic cells.
© 2008 Robert J. Robbins. All rights reserved. ![]()
Interference phenomena are yet being studied today, and enquiry has shown that interference can human activity over extremely large distances of the genome. For example, Kenneth J. Hillers and Anne M. Villeneuve recently demonstrated that in Caenorhabditis elegans , interference can actually occur over half the genome of the organism. They demonstrated this past fusing multiple chromosomes together and observing that crossovers however occurred a single time (Hillers & Villeneuve, 2003).
Complete and Incomplete Linkage
When Sturtevant drew his chromosomal map, he placed the C and O genes at the aforementioned location because they were always inherited together (Figure 3; Sturtevant, 1913). Genes that are so close together on a chromosome that they are always inherited as a single unit show a relationship referred to equally complete linkage. In fact, two genes that are completely linked tin can but exist differentiated as dissever genes when a mutation occurs in one of them. In that location is no other style to identify genes with complete linkage from unmarried genes that show multiple phenotypes.
On the other hand, the phenomenon known as incomplete linkage occurs when ii genes show linkage with a recombination level greater than 0% and less than l%. In incomplete linkage, all expected types of gametes are formed, but the recombinant gametes occur less oft than the parental gametes.
In addition, if two genes are on the same chromosome and are far plenty apart that they undergo recombination at least fifty% of the fourth dimension, the genes are independently assorting and practice not evidence linkage. Genes independently assort at a distance of 50 cM or more apart. This ways that no statistical test would allow researchers to measure linkage.
Finally, linked genes that do not independently assort bear witness statistical linkage. Statistical linkage is detected as deviation from independent assortment that favors the parental gametes. Syntenic genes are genes that are physically located on the same chromosome, whether or not the genes themselves exhibit linkage (Passarge et al., 1999). Therefore, all linked genes are syntenic, but non all syntenic genes bear witness genetic linkage.
References and Recommended Reading
Blixt, S. Why didn't Gregor Mendel find linkage? Nature 256, 206 (1975) doi:10.1038/256206a0 (link to article)
Bridges, C. B. Salivary chromosome maps with a central to the banding of the chromosomes of Drosophila melanogaster. Periodical of Heredity 26, 60–64 (1935)
———. A revised map of the salivary gland X chromosome. Journal of Heredity 29, 1113 (1938)
Hillers, One thousand., & Villeneuve, A. Chromosome-wide control of meiotic crossing over in C. elegans. Current Biology 13, 1641–1647 (2003) doi:10.1016/j.cub.2003.08.026
Morgan, T. H. Random segregation versus coupling in Mendelian inheritance. Science 34, 384 (1911)
Passarge, E., et al. Incorrect use of the term "synteny." Nature Genetics 23, 387 (1999) (link to article)
Pierce, B. Genetics: A Conceptual Approach. (New York, W. H. Freeman & Co., 2005)
Punnett, R. C. Linkage in the sweet pea (Lathyrus odoratus). Journal of Genetics thirteen, 101–123 (1923)
———. Linkage groups and chromosome number in Lathyrus. Proceedings of the Purple Society of London: Serial B, Containing Papers of a Biological Character 102 236–238. (1927)
Robbins, R. J. Introduction to sexual practice-limited inheritance in Drosophila. Electronic Scholarly Publishing Foundations of Classical Genetics Project. http://www.esp.org/foundations/genetics/classical/thm-10a.pdf (2000) (accessed May xix, 2008)
Sturtevant, A. H. The linear arrangement of half-dozen sex-linked factors in Drosophila, as shown past their mode of clan. Periodical of Experimental Zoology 14, 43–59 (1913)
Weiner, J. Time, Love, Memory: A Great Biologist and His Quest for the Origins of Behavior (New York, Random House, 1999)
Source: http://www.nature.com/scitable/topicpage/thomas-hunt-morgan-genetic-recombination-and-gene-496
0 Response to "To Determine the Order of Genes on a Chromosome, One Can Perform a"
Enregistrer un commentaire